Преимплантационное генетическое тестирование как основание для уточнения кариотипа родителей. Клинические случаи
Елизавета Мусатова¹,²*, Константин Благодатских¹, Надежда Шилова², Жанна Маркова², Марина Минженкова², Владимир Каймонов¹, Екатерина Померанцева¹, Артур Исаев¹
¹Центр генетики и репродуктивной медицины «ГЕНЕТИКО» ПАО
²Медико-генетический научный центр (Москва, РФ)
Абстракт
Цель: в статье приведено описание нескольких клинических случаев, когда результаты преимплантационного генетического тестирования (ПГТ) на хромосомные аномалии позволили заподозрить родительскую хромосомную перестройку или уточнить особенности кариотипа родителя.
Методы: ПГТ на хромосомные аномалии проводилось на образцах биопсии трофэктодермы эмбрионов, полученных методом экстракорпорального оплодотворения (ЭКО). ПГТ было выполнено с использованием сравнительной геномной гибридизации на чипах (aCGH) и секвенирования нового поколения (NGS). Уточнение кариотипов родителей проводилось с помощью стандартного цитогенетического исследования, FISH-анализа
Результаты: Во всех четырех случаях проведения ПГТ при анализе хромосомных профилей образцов трофэктодермы наблюдался неслучайный паттерн хромосомного дисбаланса. Хромосомные аномалии, выявленные в эмбрионах, позволили предположить наличие различных хромосомных перестроек в кариотипах родителей, а именно реципрокных транслокаций (случаи i, ii и iv) и сложной хромосомной перестройки (случай iii). Последующий анализ кариотипов пациентов позволил выявить инсерцию в одном случае и реципрокные транслокации в трех других. На момент тестирования эмбрионов две пары не имели информации о своих кариотипах (случаи i и ii), в то время как в случае iv было известно, что у пациентки нормальный женский кариотип, однако результаты ПГТ позволили выявить материнскую реципрокную транслокацию. В случае iii было известно, что у пациентки реципрокная транслокация, однако результаты PGT послужили основой для уточнения ее кариотипа, в результате чего была выявлена сложная хромосомная перестройка.
Заключение: Результаты преимплантационного генетического тестирования на анеуплоидии могут служить основой для уточнения кариотипов родителей. При анализе результатов ПГТ крайне важно обращать внимание на наличие закономерностей в наблюдаемом хромосомном дисбалансе, что может повлиять на ход медико-генетического консультирования пациентов и рекомендации врача по итогам проведенного протокола ЭКО с ПГТ.
DOI 10.54235/27382737-2024.v4.2-106
INTRODUCTION
Preimplantation genetic testing for aneuploidy (PGT-A) allows to select embryos with the best potential to further development and to increase the IVF efficiency. Chromosomal abnormalities are the most common genetic cause of infertility (Guo et al., 2016). The most frequent structural chromosome rearrangements in the general population and patients with infertility are autosomal reciprocal translocations (De Sutter et al., 2012). Chromosomal imbalance is the main cause of unsuccessful outcomes of embryo transfer: the implantation failures and miscarriages (Montag et al., 1997; Margalioth et al., 2006). Researchers have noted the increased chromosomal structural abnormalities frequency in patients with repeated implantation failures (Raziel et al., 2002; Coughlan et al., 2013).
Preimplantation testing of 23 chromosome pairs by methods based on comparative genomic hybridization, SNP analysis, or next generation sequencing (NGS) allow to reveal aneuploidy as well as segmental gains or losses within the resolution of the method. The comprehensive chromosome screening using methods capable of detecting segmental rearrangements allow to reveal random chromosomal imbalance in embryos at the preimplantational stage of development and also to detect the segregation of parental chromosome rearrangement in embryos. As a rule the balanced chromosome rearrangements carriers have no specific phenotypic features; however they have an increased unbalanced gametes formation risk. Thus, if one partner in the couple has a chromosome rearrangement there is an increased chromosomal abnormalities risk in embryos. In the case when one partner is a carrier of translocation, there is an increased risk of forming gametes having duplications and deletions of chromosome regions corresponding to the translocated regions.
The PGT results of chromosomal changes in embryos should correspond to parental karyotypes. Occasionally, the PGT findings in embryos make the patient karyotype questionable. We report four cases of chromosomal abnormalities revealed in embryos which allowed us to suspect different chromosomal rearrangements in parental karyotypes.
CASES PRESENTATION
Case i. The couple was directed to IVF-PGT due to the advanced maternal age and repeated implantation failures. One IVF cycle was performed and five embryos were biopsied for comprehensive chromosomal screening using aCGH (24sure, Illumina). There was no information about the karyotype of the patient.
Case ii. The PGT-A indication for the couple was the maternal age. One IVF cycle was performed and four embryos were biopsied for PGT by NGS (Veriseq, Illumina).
Case iii. The couple was directed to IVF-PGT due to the maternal balanced reciprocal translocation. The same result of maternal karyotype analysis performed in two independent laboratories was obtained: 46,ХХ,t(4;13)(p14;q34). Two IVF cycles were performed and nine embryos were biopsied for PGT by aCGH (24sure+, Illumina).
Case iv. Due to infertility a standard peripheral blood lymphocyte culture cytogenetic study was performed for both partners and the obtained results were 46,XX and 46,XY,inv(10)(p11.2q22). The PGT indication for the couple was paternal pericentric inversion of chromosome 10. Two IVF cycles were performed and four embryos were biopsied for PGT by NGS (Veriseq, Illumina).
MATERIALS AND METHODS
PGT-A of embryos obtained by in vitro fertilization (IVF) were performed by microarray comparative genomic hybridization (aCGH) using microarrays with different resolution (24sure+/24sure, Illumina) (cases i and iii) and by NGS (VeriSeq, Illumina) (case ii and iv). The choice of microarray resolution was determined by the indication for preimplantation testing. The trophectoderm biopsy was performed at 5-6 day of embryo development. The parental karyotype detection was performed by fluorescence in situ hybridization (FISH) (cases ii, iii and iv) and parental karyotype analysis was performed by standard cytogenetic study of peripheral blood lymphocyte culture (case i). The informed consent was obtained from all individuals included in the study.
RESULTS
Case i. The couple was directed to IVF-PGT due to the advanced maternal age and repeated implantation failures. One IVF cycle was performed and five embryos were biopsied for comprehensive chromosomal screening using aCGH (24sure, Illumina). There was no information about the karyotype of the patient. The PGT results for chromosomal abnormalities are shown in Fig. 1 and Supplemental table 1. In the case of samples T8 and T11, fertilization was achieved using the donor sperm and random aneuploidies were detected in these two samples. In the remaining three samples, segmental rearrangements were detected as duplications and deletions of chromosomes 11 and 22 regions. These PGT results allowed to suspect that one partner in the couple appeared to be the reciprocal translocation carrier. A standard cytogenetic study of peripheral blood lymphocyte culture of both partners was performed. The couple karyotypes were identified as 46,ХХ and 46,ХY,t(11;22)(q23.3;q13). Thus, the paternal reciprocal translocation between chromosomes 11 and 22 was detected.
Case ii. The PGT-A indication for the couple was the maternal age. One IVF cycle was performed and four embryos were biopsied for PGT by NGS. The preimplantation testing results for chromosomal abnormalities are shown in Fig. 2А and Supplemental table 2. All four samples contained the repeated segmental arrangements as chromosomes 10 and 12 distal regions deletions/duplications. A nonrandom pattern of segmental losses and gains allowed one to suspect that one partner in the couple appeared to be the reciprocal translocation carrier. The patients’ karyotypes were normal: a standard cytogenetic study of peripheral blood lymphocyte culture was performed for both partners and the results obtained were 46,XX and 46,XY. FISH analysis was recommended to the couple. The reciprocal translocation between chromosomes 10 and 12 was detected by maternal cultured lymphocytes FISH analysis (Fig.2В).
Case iii. The couple was directed to IVF-PGT due to the maternal balanced reciprocal translocation. The same result of maternal karyotype analysis performed in two independent laboratories was obtained: 46,ХХ,t(4;13)(p14;q34). Two IVF cycles were performed and nine embryos were biopsied for PGT by aCGH (24sure+, Illumina). The PGT results are shown in Supplemental table 3. Seven samples out of nine were abnormal while all abnormal samples contained segmental deletions or duplications of the chromosome 4 interstitial region. An analysis of chromosomal profiles in embryos has revealed chromosome 4 segmental rearrangements with the malsegregation pattern which is not typical for translocation since the quantitative imbalance was observed in the chromosome 4 interstitial region but not in the telomeric region (Fig. 3). In addition, a small number of normal embryos were observed in both IVF cycles. These findings allowed to question the karyotype structure accuracy. The FISH analysis was performed using maternal cultured lymphocytes which confirmed the fact that the female patient had complex chromosomal rearrangement: the chromosome 4 p-arm part interchromosomal insertion into chromosome 13 q-arm and the chromosome 4 pericentric inversion (Fig. 4).
Case iv. Due to infertility a standard peripheral blood lymphocyte culture cytogenetic study was performed for both partners and the obtained results were 46,XX and 46,XY,inv(10)(p11.2q22). The PGT indication for the couple was paternal pericentric inversion of chromosome 10. Two IVF cycles were performed and four embryos were biopsied for PGT by NGS. All four samples were abnormal and three of four samples contained the repeated segmental arrangements as chromosomes 2 and 12 distal regions deletions/duplications. Chromosome 2 segmental abnormalities were as small as 3,3 Mb and didn’t conclude in clinical results (Supplemental table 4). A nonrandom pattern of segmental losses and gains allowed to suspect that one partner in the couple appeared to be the reciprocal translocation carrier. FISH analysis was recommended to the couple. The maternal cultured lymphocytes reciprocal translocation between chromosomes 2 and 12 was detected by FISH analysis (Fig.5).
DISCUSSION
The presented cases differ by PGT indications, initial information about parental karyotypes, and methods of embryo analysis; however, they were combined by a key role of the embryo genetic testing performed. In all these cases, the use of PGT-A not only allowed to obtain information about the embryos possessing the highest potential for further development but also determined the necessity of patient karyotype study. The embryos that turned out to be unusable for the embryo transfer due to the chromosomal imbalance provided valuable information about the possible parental chromosomal rearrangement and its segregation. The nonrandom character of chromosomal abnormalities in the embryo samples studied by PGT should be noticed since it can be the indication for a patient karyotype analysis. In addition to the mere fact of chromosomal imbalance nonrandom character in the studied embryo samples, it is important to notice the suggested segregation patterns of the parental chromosomal rearrangements. In the case iii, before performing the IVF cycle and PGT, it was known that the female partner has the structural chromosomal rearrangement. Nevertheless, the segregation pattern revealed by PGT was untypical for reciprocal translocation, which allowed to establish the correct karyotype and to define the genetic risks, as well as to explain the unexpectedly high portion of abnormal embryos for the couple whose age was less than 35 years since insertions bear the highest reproductive risk compared with translocations.
PGT is not only the precise method for aneuploidy testing but also has a potential to improve karyotype diagnostics for the couple. Some patients enter the IVF programs having no information about their karyotypes. However, even in the case when parental karyotypes were known and no pathology was revealed, we can not exclude the occurrence of submicroscopic structural rearrangements. NGS and aCGH methods that are used for PGT to detect chromosomal abnormalities have the resolving power that allows to reveal the quantitative chromosomal abnormalities from the parental chromosomal rearrangements which are too small to be detected by the standard cytogenetic analysis. It is extremely important to take into account the embryo chromosomal rearrangements pattern obtained by PGT. In some cases, the PGT results can serve as a basis for karyotyping patients (Sundheimer et al., 2018). Otherwise, the karyotype revision may be recommended (Frumkin et al., 2017). In the case, when the cytogenetic diagnosis for the couple is already established, the embryo chromosomal profiles should correspond to the parental karyotypes. The PGT findings can help to identify the carriers of chromosomal rearrangements among the IVF patients. Thus, the correct information about the patient karyotype can allow us to identify offspring genetic risks and to explain the couple’s infertility cause.
References
- Coughlan, C., Yuan, X., Nafee, T., Yan, J., Mariee, N., and Li, T. (2013). The clinical characteristics of women with recurrent implantation failure. J. Obstet. Gynaecol. 33, 494–498. doi:10.3109/01443615.2013.782280.
- De Sutter, P., Stadhouders, R., Dutré, M., Gerris, J., and Dhont, M. (2012). Prevalence of chromosomal abnormalities and timing of karyotype analysis in patients with recurrent implantation failure (RIF) following assisted reproduction. Facts Views Vis. ObGyn 4, 59–65.
- Frumkin, T., Peleg, S., Gold, V., Reches, A., Asaf, S., Azem, F., et al. (2017). Complex chromosomal rearrangement—a lesson learned from PGS. J. Assist. Reprod. Genet. 34, 1095–1100. doi:10.1007/s10815-017-0954-y.
- Guo, K. M., Wu, B., Wang, H. B., and Tian, R. H. (2016). Reproductive outcome of male carriers of chromosomal abnormalities: multidisciplinary approach for genetic counseling and its implications. Genet. Mol. Res. 15. doi:10.4238/gmr15048963.
- Margalioth, E. J., Ben-Chetrit, A., Gal, M., and Eldar-Geva, T. (2006). Investigation and treatment of repeated implantation failure following IVF-ET. Hum. Reprod. 21, 3036–3043. doi:10.1093/humrep/del305.
- Montag, M., van der Ven, K., Ved, S., Schmutzler, A., Prietl, G., Krebs, D., et al. (1997). Success of intracytoplasmic sperm injection in couples with male and/or female chromosome aberrations. Hum. Reprod. 12, 2635–2640. doi:10.1093/humrep/12.12.2635.
- Raziel, A., Friedler, S., Schachter, M., Kasterstein, E., Strassburger, D., and Ron-El, R. (2002). Increased frequency of female partner chromosomal abnormalities in patients with high-order implantation failure after in vitro fertilization. Fertil. Steril. 78, 515–519. doi:10.1016/S0015-0282(02)03298-3.
- Sundheimer, L. W., Liu, L., Buyalos, R. P., Hubert, G., Al-Safi, Z., and Shamonki, M. (2018). Diagnosis of parental balanced reciprocal translocations by trophectoderm biopsy and comprehensive chromosomal screening. J. Assist. Reprod. Genet. 35, 165–169. doi:10.1007/s10815-017-1042-z.
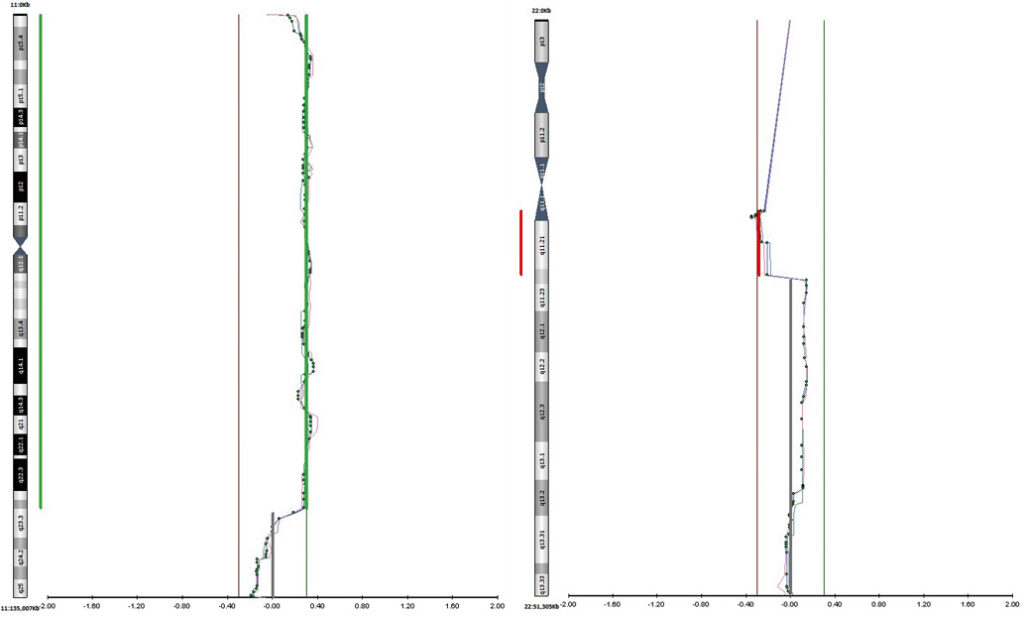
Figure 1. Chromosomes 11 and 22 profiles for trophectoderm biopsy sample T1 (case i).
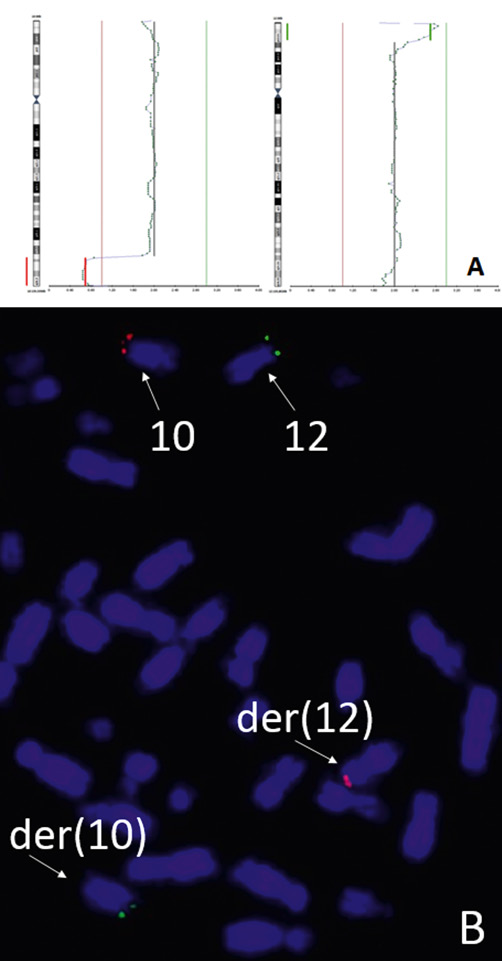
Figure 2. Case ii. The PGT results for chromosomes 10 and 12 of trophectoderm biopsy sample T2 and the FISH analysis using maternal cultured lymphocytes: (A), chromosomes 10 and 12 profiles for trophectoderm biopsy sample T2; (B), subtelomeric probes for q-arm (red) of chromosome 10 and p-arm (green) of chromosome 12 of maternal cultured lymphocytes (the FISH analysis).
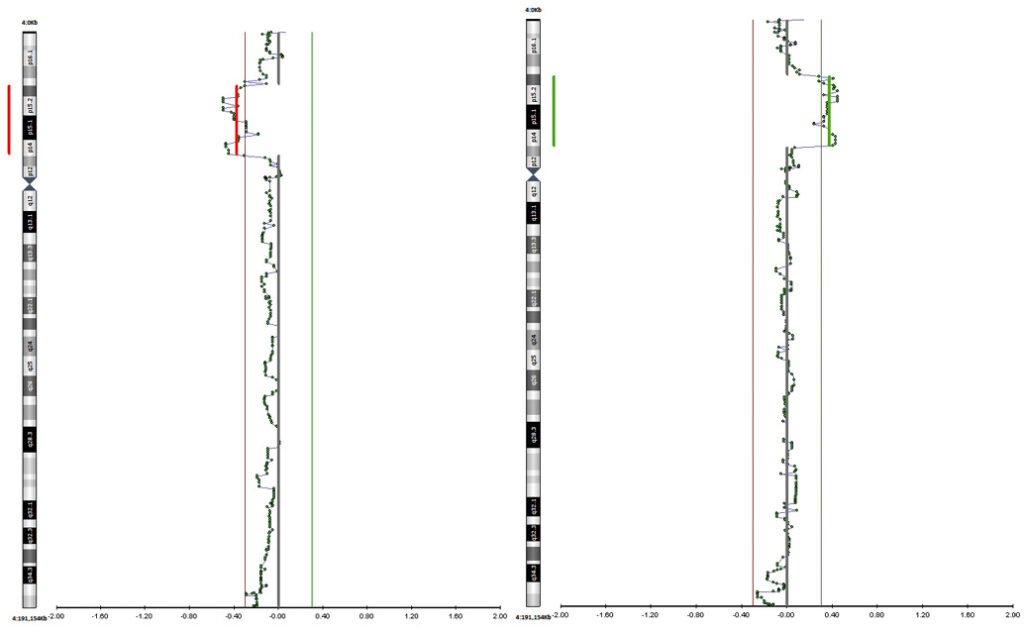
Figure 3. Chromosomе 4 profile for samples T1 and T2 from the IVF cycle N1 (case iii).
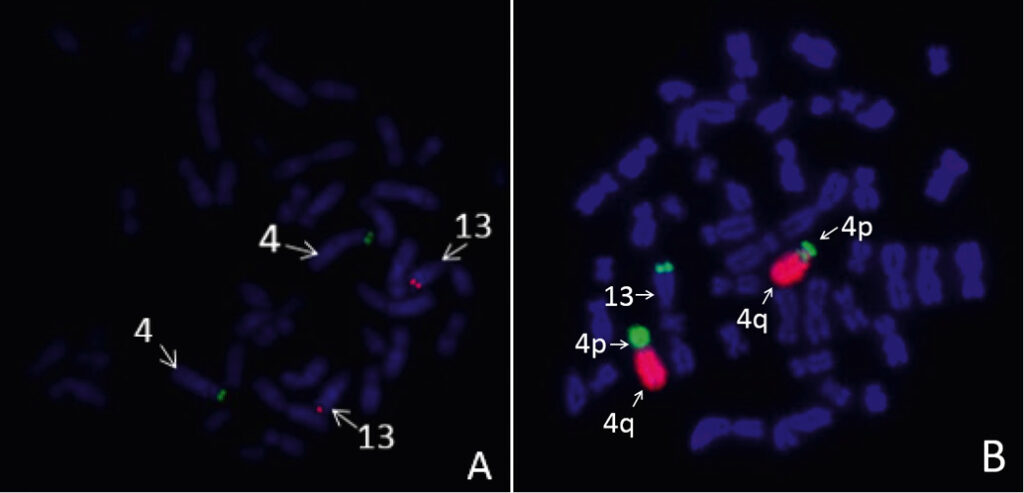
Figure 4. Results of the FISH analysis using maternal cultured lymphocytes (case iii). A, subtelomeric probes for p-arm (green) of chromosome 4 and q-arm (red) of chromosome 13. B, arm-specific probes for q-arm (red) and p-arm (green) of chromosome 4.
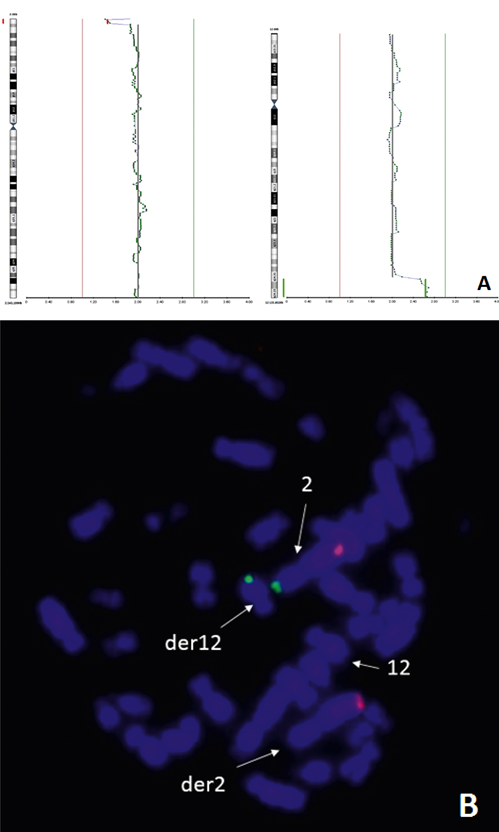
Figure 5. Case iv. The PGT results for chromosomes 2 and 12 of trophectoderm biopsy sample and the FISH analysis using maternal cultured lymphocytes: A, chromosomes 2 and 12 profiles for trophectoderm biopsy sample; B, subtelomeric probes for q-arm (red) and p-arm (green) chromosome 2 on maternal cultured lymphocytes.